Background
In the first week, we learnt some background information about RNA-seq technology. Next-generation sequencing is applied by RNA-seq technology, which is also called whole transcriptome shotgun sequencing, to determine if some specific RNA for differentiated gene expression is present in the genomic DNA and what might be the number of it in the sample at a specific time. To get the potential modified parts identified, the whole gene will be tested and it is cut into pieces so that they can be more easily analyzed. Normally, the size of each RNA fragment is always about 80-110 bp. After the RNA chain is cut into pieces, the entire known gene database will be to get compared with them to identify the modified parts that have effects on the change of functional expression. With the data that we got, RNA-seq technology was used to determine the differences between two groups of samples, “58” and “60”.
Importing transcript-level estimates
As the samples were saved, we needed to get them imported into R and Salmon and tximport packages to were used.
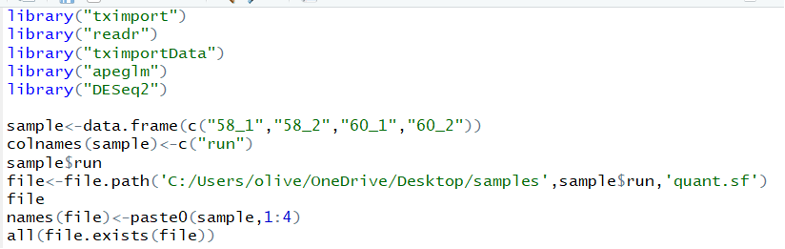
After importing these four samples, “58_1”, “58_2”, “60_1”, and “60_2”, a direction was created to contain them and it was check with typing the specific code to determine if the system would turn “TRUE” to the next step.
Incorporating the transcript-level estimates into gene-level estimates
In this step, TxDb package, TxDb.Mmusculus.UCSC.mm10.knownGene, was inserted to get the transcript-level estimates transferred to the gene-level estimates. One data frame, which is called tx2geen, was created and contained two columns, transcript ID and gene ID. The entire known gene database was applied to get the transcript-level estimates compared with it so that it can be checked if the gene ID present in the tested sample exists in the database. And then a direction, called txdb, was created to contain the the gene database of Mus musculus known gene to take the information.
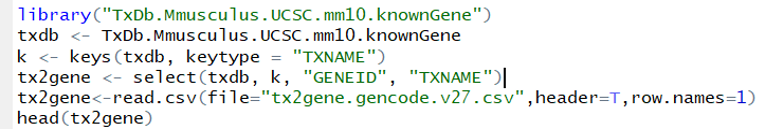
The ones shown in the columns are transcript names and gene ID where they belong to.
Differential Expression Analysis
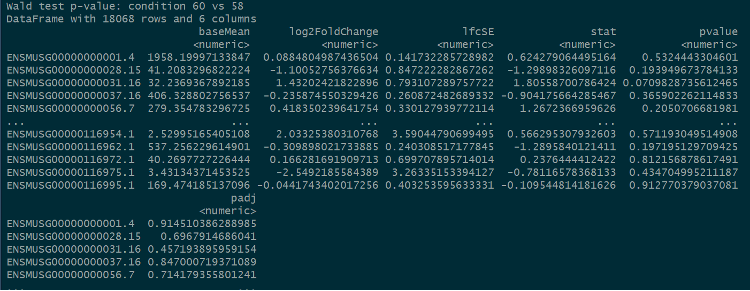
For the next step, the gene-level data would be used to do the further analysis with the package, “DESeq2”, to determine the log2FoldChange and adjusted p-values.

Two conditions, “58” and “60”, were created to contain there four samples and another data frame was created by the “DESeq2” function and “result” function.
Quality control
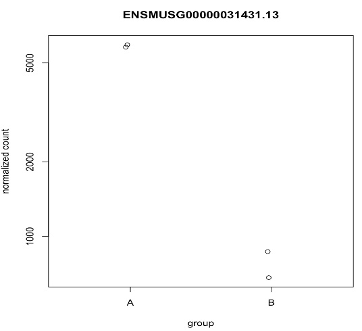
Plots counts

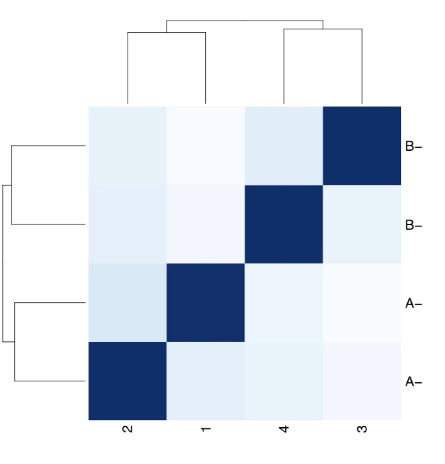
Heatmap
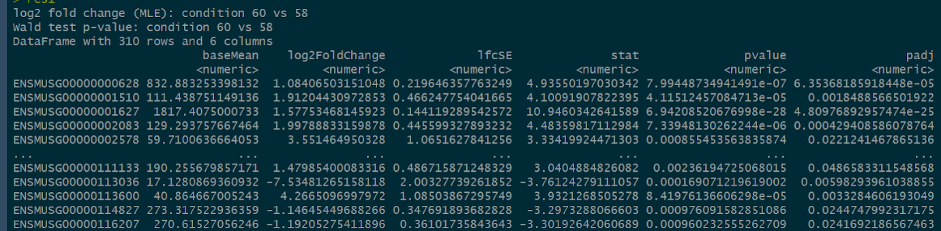
Final gene list
To generate the final gene list, there were two conditions created to extract the useful information, p-value is less than 0.05 and log2FoldChange is greater than 1.
Functions analysis
With the use of GO analysis and application of Generatio, we tried to identify the differences between the expressions performed by the genen products of “58” and “60” based on some areas, such as epidermal cell differentiation, skin development, and so on. From the differences, we can get the further deduction and prove it - if there are stem cells added to the epithelial tissues, which ascribes to the remarkable differentiations of related cells.
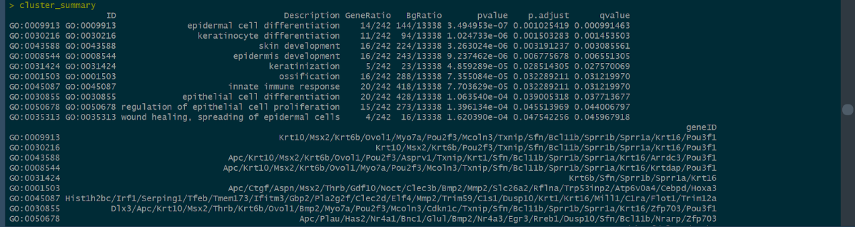
The picture below shows how many genes are related to epidermal cell differentiation, keratinocyte differentiation, skin development, and keratinization with the identification of the size of spheres.
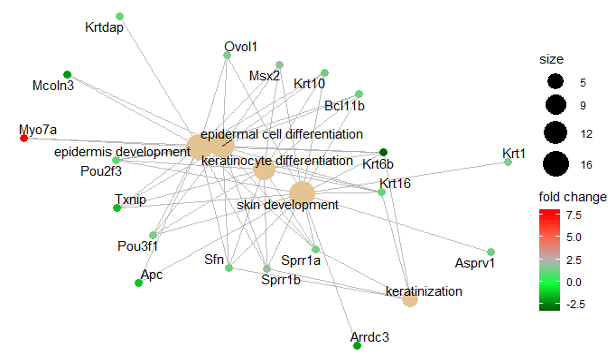
The picture below shows the sizes of spheres, which represents the number of genes related to it, and ten most significant terms check in the experiments with the collected data. If the adjusted p-value ranges from 0.01 to 0.04 and the color of spheres ranges from red to purple. If the color is brighter red, the adjusted p-value is greater.

The genes involved in the experiment is shown below
Conclusion
Two groups of data were compared to identify the differences with different methods and packages in R. Additionally, the results show that the genes checked in the experiment are related to the epidermal cell differentiation, skin development, epithelial cell differentiation, and so on.
In the third week, we were assigned to do the presentation about our interested topics and do some other introductions about the future direction of our topics. The topic that I chose is the detection of dopamine with carbon fiber microelectrodes coated with gold nanoparticles.
In the presentation, I introduced two main areas that produced dopamine, which is responsible for physical movements and people’s cognitive abilities and behaviors, and the diseases that are related to the amount of dopamine are Parkinson’s disease and schizophrenic. Besides, I showed the definition of nanoparticles and their applications in medical, electric and environment areas as nanoparticles’ physical, chemical, and biological properties will get changed when they get transferred from bulk materials to nanoscale. Additionally, as we used protein-assisted method in the production of gold nanoparticles, so we determined the factors that might affect the efficient production, molecular ratio of gold chloride and amino acids and pH range. The methods applied in the experiments were also introduced in the presentation, such as electrodeposition, cutting the capillary contained with single carbon fiber into carbon fiber microelectrodes, and so on.
The most import point in the presentation is the future direction. To make progress in my research project, my partner and I will make the silver nanoparticles for the future detection of dopamine as we assume that the reduction silver ions, positive one, need less time and less current than the reduction of gold ions, positive three. What’s more, as silver nanoparticles are toxic to human bodies, I will try to find nanoparticles composed of other metals that will not have effects on human’s health. In addition to this, as Parkinson’s disease is caused by the loss of dopamine and the degeneration of neurons, I also want to do the further experiments and find resources to check if it will be possible to achieve the production of neurons with neurons stem cells in the laboratory with the methods of cell culture. Besides, as nanoparticles can be used to transfer drugs to specific cells to achieve the treatment. Therefore, I also want to prove if it can be possible to have the nanoparticles transferred the produced neurons into the brain and achieve the accurate implantation of neurons to get this disease totally treated. In the fourth week, I will try find to more articles to learn about the process of transferring drugs to specific cells with nanoparticles and other thoughts and find data that can determine the implementability of the hypothesis to design the future experiment.



